The joker package provides a comprehensive set of
features for probabilities and mathematical statistics. It extends the
range of available distribution families and facilitates the computation
of key parametric quantities, such as moments and information-theoretic
measures. The main focus of the package is parameter estimation through
maximum likelihood and moment-based methods under an intuitive and
efficient coding framework.
Introduction
Package joker is designed to cover a broad collection of
distribution families, extending the functionalities of the
stats package to support new families, parametric quantity
computation and parameter estimation. All package features are available
both in a stats-like syntax for entry-level users, and in
an S4 object-oriented programming system for more
experienced ones.
The joker S4 Distribution System
Probability Distributions
In the joker OOP system, each distribution has a
respective S4 class. The following table shows the
distributions available in the package, along with their class names.
All of them are subclasses of the Distribution
S4 class.
library(knitr)
kable(
data.frame(
Distribution = c("Bernoulli", "Beta", "Binomial", "Categorical", "Cauchy",
"Chi-Square", "Dirichlet", "Fisher", "Gamma", "Geometric"),
Class_Name = c("Bern", "Beta", "Binom", "Cat", "Cauchy", "Chisq", "Dir",
"Fisher", "Gam", "Geom"),
Distribution2 = c("Laplace", "Log-Normal", "Multivariate Gamma",
"Multinomial", "Negative Binomial", "Normal", "Poisson",
"Student", "Uniform", "Weibull"),
Class_Name2 = c("Laplace", "Lnorm", "Multigam", "Multinom", "Nbinom",
"Norm", "Pois", "Stud", "Unif", "Weib")
),
col.names = c("Distribution", "Class Name", "Distribution", "Class Name"),
caption = "Overview of the distributions implemented in the `joker` package,
along with their respective class names."
)| Distribution | Class Name | Distribution | Class Name |
|---|---|---|---|
| Bernoulli | Bern | Laplace | Laplace |
| Beta | Beta | Log-Normal | Lnorm |
| Binomial | Binom | Multivariate Gamma | Multigam |
| Categorical | Cat | Multinomial | Multinom |
| Cauchy | Cauchy | Negative Binomial | Nbinom |
| Chi-Square | Chisq | Normal | Norm |
| Dirichlet | Dir | Poisson | Pois |
| Fisher | Fisher | Student | Stud |
| Gamma | Gam | Uniform | Unif |
| Geometric | Geom | Weibull | Weib |
Defining an object from the desired distribution class is
straightforward, as seen in the following example. The parameter names,
which are generally identical to the ones defined in the
stats package, can be omitted.
shape1 <- 1
shape2 <- 2
D <- Beta(shape1, shape2)Having defined the distribution object D, the
d()-p()-q()-r()
functions can be used, as shown in the following example, comparing
against the stats syntax.
d(D, 0.5)
> [1] 1
dbeta(0.5, shape1, shape2)
> [1] 1
p(D, 0.5)
> [1] 0.75
pbeta(0.5, shape1, shape2)
> [1] 0.75
qn(D, 0.75)
> [1] 0.5
qbeta(0.75, shape1, shape2)
> [1] 0.5
r(D, 2)
> [1] 0.2494058 0.3353119
rbeta(2, shape1, shape2)
> [1] 0.15414668 0.06689389Alternatively, if only the distribution argument is supplied, the methods behave as functionals (i.e. they return a function). This behavior offers enhanced functionality such as:
Technical Detail: The quantile function is called
qn() rather than the more intuitive q(). The
reason behind this choice lies in the RStudio IDE
(Integrated Development Environment), which overrides the method
selection process of the base function q()
used to quit an R session, i.e. a function named
q() always ends the session. In order to avoid this
unpleasant behavior, the name qn() was chosen instead.
Parametric Quantities of Interest
The joker package contains a set of methods that
calculate the theoretical moments (expectation, variance and standard
deviation, skewness, excess kurtosis) and other important parametric
functions (median, mode, entropy, Fisher information) of a distribution.
Alternatively, the moments() function automatically finds
the available methods for a given distribution and returns all of the
results in a list.
mean(D)
> [1] 0.3333333
median(D)
> [1] 0.2928932
mode(D)
> [1] 0
var(D)
> [1] 0.05555556
sd(D)
> [1] 0.2357023
skew(D)
> [1] 0.5656854
kurt(D)
> [1] 0.2333333
entro(D)
> [1] -0.1931472
finf(D)
> shape1 shape2
> shape1 1.2500000 -0.3949341
> shape2 -0.3949341 0.2500000Technical Detail: Only the function-distribution
combinations that are theoretically defined are available; for example,
while var() is available for all distributions,
sd() is available only for the univariate ones. In case the
result is not unique, a predetermined value is returned with a warning.
The following example illustrates this in the case of
,
i.e. a uniform distribution for which every value in the
interval is a mode.
Parameter Estimation
The joker package includes a number of options when it
comes to parameter estimation. In order to illustrate these
alternatives, a random sample is generated from the Beta
distribution.
Estimation Methods
The package covers three major estimation methods: maximum likelihood estimation (MLE), moment estimation (ME), and score-adjusted moment estimation (SAME).
In order to perform parameter estimation, a new
e<name>() member is added to the
d()-p()-q()-r()
family, following the standard stats name convention. These
e<name>() functions take two arguments, the
observations x (an atomic vector for univariate or a matrix
for multivariate distributions) and the type of estimation
method to use.
ebeta(x, type = "mle")
> $shape1
> [1] 1.066968
>
> $shape2
> [1] 2.466715
ebeta(x, type = "me")
> $shape1
> [1] 1.074511
>
> $shape2
> [1] 2.469756
ebeta(x, type = "same")
> $shape1
> [1] 1.067768
>
> $shape2
> [1] 2.454257A general function called e() is also implemented,
covering all distributions and estimators.
Dominant estimation methods such as the mle(),
me(), and same() are also available as
S4 generics.
mle(D, x)
> $shape1
> [1] 1.066968
>
> $shape2
> [1] 2.466715
me(D, x)
> $shape1
> [1] 1.074511
>
> $shape2
> [1] 2.469756
same(D, x)
> $shape1
> [1] 1.067768
>
> $shape2
> [1] 2.454257Technical Detail: It is important to note that the
S4 methods also accept a character for the distribution.
The name should be the same as the S4 distribution
generator, case ignored.
Log-likelihood
Likewise, log-likelihood functions are available in two versions, the
distribution specific one, e.g. llbeta(), and the
ll() S4 generic one.
In some distribution families like beta and gamma, the MLE cannot be
explicitly derived and numerical optimization algorithms have to be
employed. Even in good scenarios, with plenty of observations
and a smooth optimization function, numerical algorithms should not be
viewed as panacea, and extra care should be taken to ensure a fast and
right convergence if possible. Two important steps are taken in
joker in this direction:
An illustrative example for the Beta distribution is shown below. Let denote the probability density function of :
where is the Gamma function. Then, the log-likelihood function, divided by the sample size , takes the form:
The score equation for is:
The score equation for is:
These two nonlinear equations must be solved numerically. However, instead of solving the above two-dimensional problem, one can see that by denoting , the two score equations can be rewritten as:
i.e. restricted to the score equation system solution space, both parameters can be expressed as a function of their sum , and therefore the log-likelihood function can be optimized with respect to :
Technical Detail: It would perhaps be more intuitive to use the score equations to express as a function of or vice versa. However, the above method can be directly generalized to the Dirichlet case and reduce the initial -dimensional problem to a unidimensional one. The same technique can be utilized for the gamma and multivariate gamma distribution families, also reducing the dimension to unity, from and respectively.
In joker, the resulting function that is inserted in the
optimization algorithm is called lloptim(), and is not to
be confused with the actual log-likelihood function ll().
The corresponding derivative is called dlloptim().
Therefore, whenever numerical computation of the MLE is required,
joker calls the optim() function with the
following arguments:
-
lloptim(), an efficient function to be optimized, -
dlloptim(), its analytically-computed derivate, - an initialization point (
numeric) or the method to use to provide one (character, e.g."me") - an optimization algorithm. By default, the L-BFGS-U is used, with lower and upper limits defined at or near the parameter space boundary.
Asymptotic Variance - Covariance Matrix
The asymptotic variance (or variance - covariance matrix for
multidimensional parameters) of the estimators are also covered in the
package by the v<name>() functions.
vbeta(shape1, shape2, type = "mle")
> shape1 shape2
> shape1 1.597168 2.523104
> shape2 2.523104 7.985838
vbeta(shape1, shape2, type = "me")
> shape1 shape2
> shape1 2.1 3.3
> shape2 3.3 9.3
vbeta(shape1, shape2, type = "same")
> shape1 shape2
> shape1 1.644934 2.539868
> shape2 2.539868 8.079736As with point estimation, the implementation is twofold, and the
general function v() covers all distributions and
estimators.
Estimation Metrics and Comparison
The different estimators of a parameter can be compared based on both
finite sample and asymptotic properties. The package includes two
functions named small_metrics() and
large_metrics() , where small and large refers to the
small sample and large sample terms that are often
used for the two cases. The former estimates the bias, variance and root
mean square error (RMSE) of the estimator with Monte Carlo simulations,
while the latter calculates the asymptotic variance - covariance matrix
(as derived by the v() functions). The resulting data
frames can be plotted with the plot() function.
To illustrate the function’s design, consider the following example from the beta distribution: We are interested in calculating the metrics (bias, variance, and RMSE) of the parameter estimators (MLE, ME, and SAME), for sample sizes 20 and 50. Specifically, we want to illustrate how these metrics change for , and (constant). The following code can do that:
D <- Beta(1, 2)
prm <- list(name = "shape1",
val = seq(1, 5, by = 0.5))
x <- small_metrics(D, prm,
obs = c(20, 50),
est = c("mle", "same", "me"),
sam = 1e3,
seed = 1)
head(x@df)
> Parameter Observations Estimator Metric Value
> 1 1.0 20 mle Bias 0.1322510
> 2 1.5 20 mle Bias 0.2486026
> 3 2.0 20 mle Bias 0.3276922
> 4 2.5 20 mle Bias 0.5215973
> 5 3.0 20 mle Bias 0.5517199
> 6 3.5 20 mle Bias 0.5381523The small_metrics() function takes the following
arguments:
-
D, the distribution object of interest, -
prm, a list that specifies how theshape1parameter values should change, -
obs, a numeric vector holding the sample sizes, -
est, a character vector specifying the estimators under comparison, -
sam, the Monte Carlo sample size to use for the metrics estimation, -
seed, a seed to be passed toset.seed()for replicability.
The resulting data frame can be passed to plot() to see
the results. The package’s plot() methods depend on
ggplot2 to provide highly-customizable graphs.
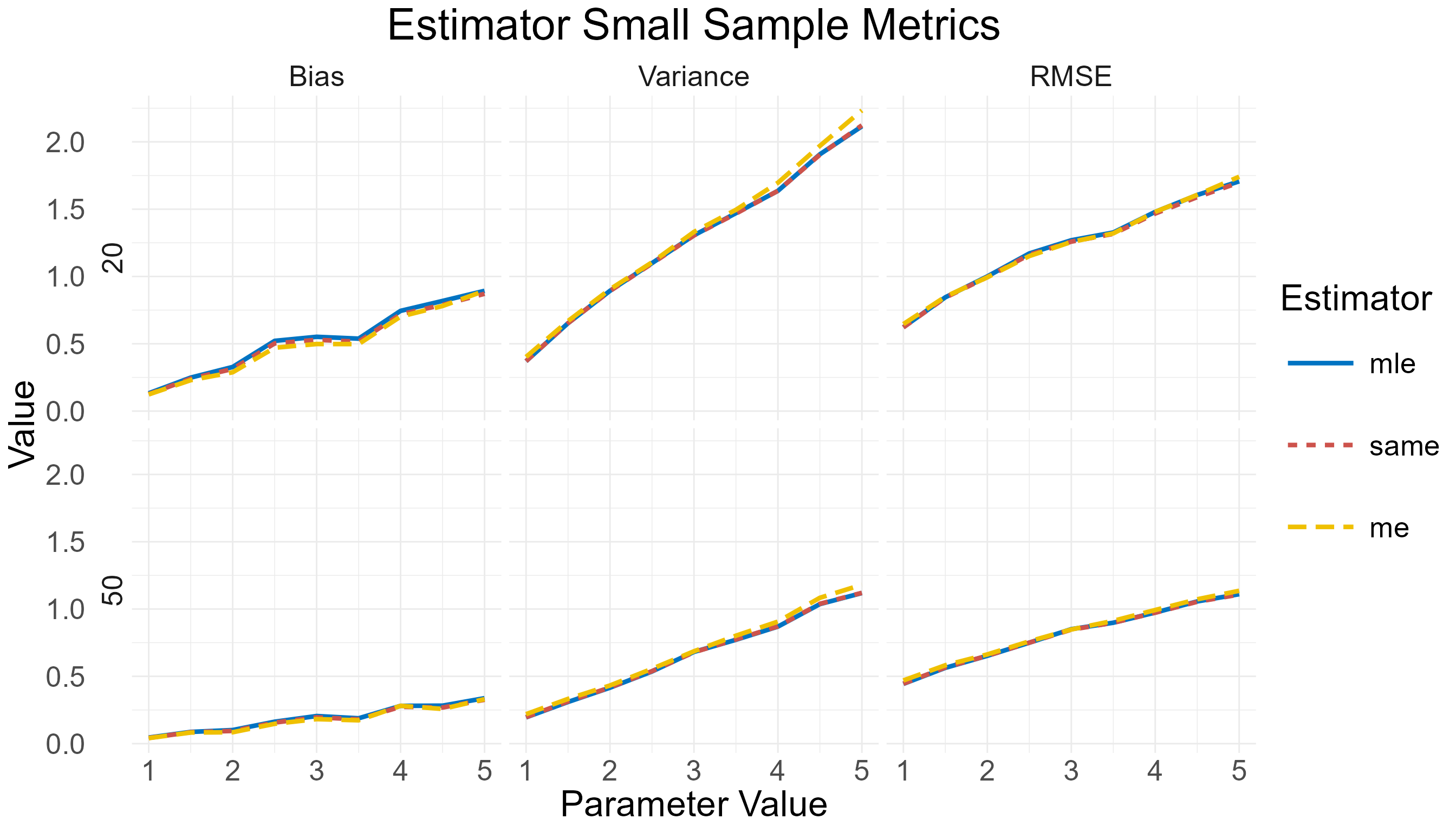
Small-sample metrics comparison for MLE, ME, and SAME of the beta distribution shape1 parameter.
Note that in some distribution families the parameter is a vector, as
is the case with the Dirichlet distribution (a multivariate
generalization of beta), which holds a single parameter vector
alpha. In these cases, the prm list can
include a third element, pos, specifying which parameter of
the vector should change:
D <- Dir(alpha = 1:4)
prm <- list(name = "alpha",
pos = 1,
val = seq(1, 5, by = 0.5))
x <- small_metrics(D, prm,
obs = c(20, 50),
est = c("mle", "same", "me"),
sam = 1e3,
seed = 1)
class(x)
> [1] "SmallMetrics"
> attr(,"package")
> [1] "joker"
head(x@df)
> Parameter Observations Estimator Metric Value
> 1 1.0 20 mle Bias 0.07759434
> 2 1.5 20 mle Bias 0.11916042
> 3 2.0 20 mle Bias 0.17488071
> 4 2.5 20 mle Bias 0.20523185
> 5 3.0 20 mle Bias 0.32907679
> 6 3.5 20 mle Bias 0.32915767The large_metrics() function design is almost identical,
except that no obs, sam, and seed
arguments are needed here. The following example illustrates the large
sample metrics for the beta distribution shape
estimators. Again, the resulting data frame can be passed to
plot().
D <- Beta(1, 2)
prm <- list(name = "shape1",
val = seq(1, 5, by = 0.1))
x <- large_metrics(D, prm,
est = c("mle", "same", "me"))
class(x)
> [1] "LargeMetrics"
> attr(,"package")
> [1] "joker"
head(x@df)
> Row Col Parameter Estimator Value
> 1 shape1 shape1 1.0 mle 1.597168
> 2 shape2 shape1 1.0 mle 2.523104
> 3 shape1 shape2 1.0 mle 2.523104
> 4 shape2 shape2 1.0 mle 7.985838
> 5 shape1 shape1 1.1 mle 1.969699
> 6 shape2 shape1 1.1 mle 2.826906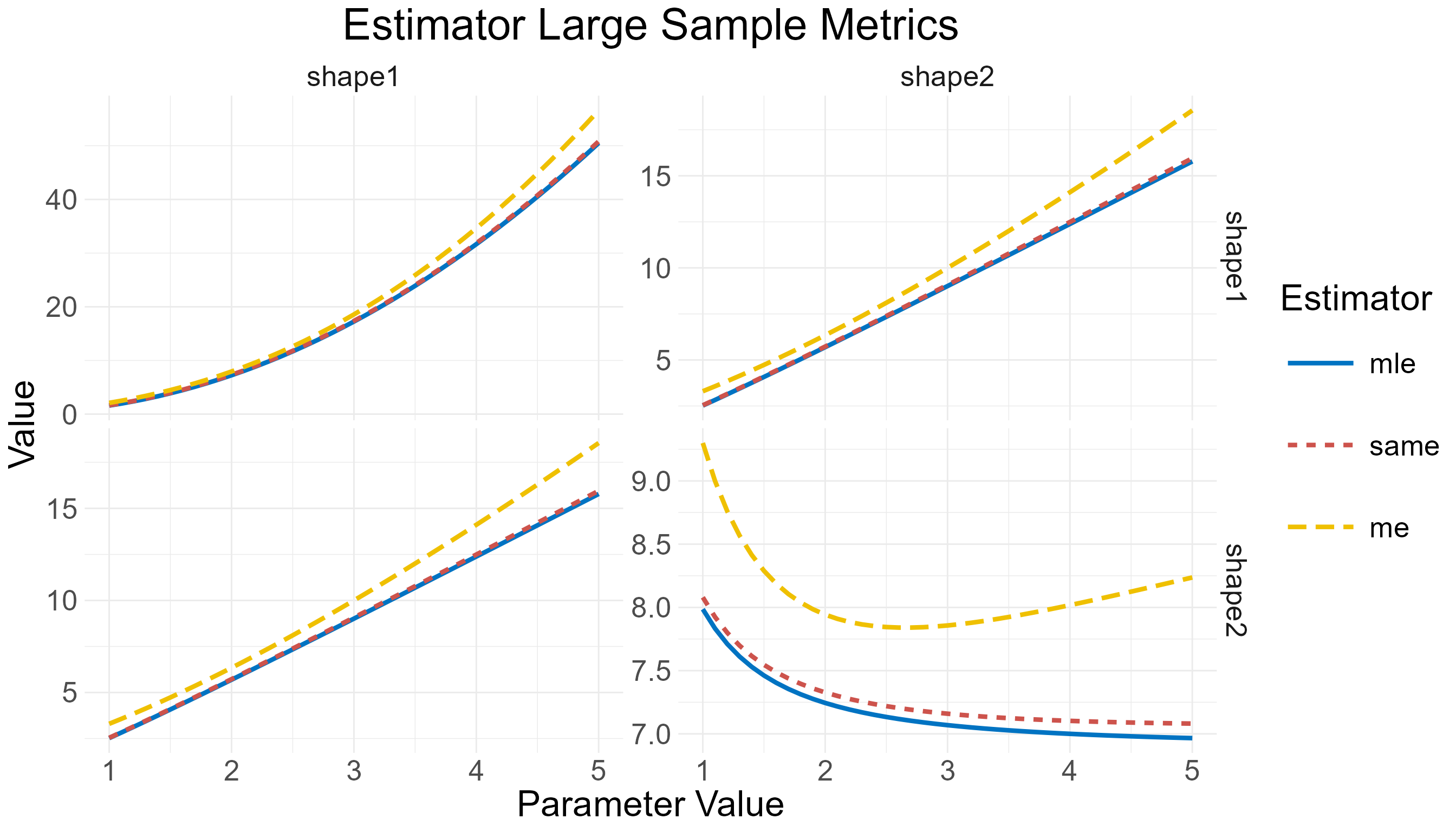
Large-sample metrics comparison for MLE, ME, and SAME of the beta distribution shape1 parameter.
Documentation and Testing
Comprehensive and clear documentation should be a central focus of
every package. In joker, all functions are extensively
documented, providing detailed descriptions of arguments, return values,
and underlying behavior. Examples are always included to illustrate
typical usage and edge cases, making it easier for users to quickly
understand and apply the functions in their workflows. In addition to
the function-level documentation, the package includes a thorough
vignette that offers a broader overview of its functionality,
demonstrates common use cases, and provides guidance on integrating the
package into larger analysis pipelines, as well as information on how to
expand its functionalities. This layered approach to documentation
ensures that users at all levels—from new adopters to experienced
developers—can make full and effective use of the package.
The joker package places a strong emphasis on rigorous
testing and quality assurance. Automated testing is implemented using
the testthat package, with a comprehensive suite of over
1,000 individual tests covering all core functionality. These tests
ensure that all features behave as expected, edge cases are handled
appropriately, and any regressions are quickly detected. Test coverage,
as measured by the covr package, exceeds 90%, reflecting a
thorough approach to exercising the codebase and providing a high level
of confidence in the package’s reliability.
To maintain consistent code quality and immediate feedback on changes, continuous integration (CI) is set up via GitHub and CircleCI. Every pull request and push to the repository triggers a full build and test cycle, ensuring that new contributions meet the package’s standards before they are merged. This automated workflow helps to detect issues early, streamlines collaboration, and supports a development process that prioritizes stability and reproducibility.
In addition to automated testing and CI, the package complies with
broader community standards and best practices. It has been validated
using rOpenSci’s pkgcheck package, which ensures adherence
to guidelines for documentation, code structure, and usability. Further
quality assurance is provided through autotest, which
automatically generates tests to explore unexpected code behaviors, and
srr (Statistical Software Review), which ensures compliance
with best practices specific to statistical software. Together, these
tools and processes help ensure the package remains robust,
maintainable, and trustworthy for users.
Defining New Classes and Methods
Of course, it is possible to be interested in a distribution family
not included in the package. It is straightforward for users to define
their own S4 class and methods. Since this paper is
addressed to both novice and experienced R users, the beta
distribution paradigm is explained in detail below:
Defining the Class
The setClass() function defines a new S4
class, i.e. the distribution of interest. The slots
argument defines the parameters and their respective class (usually
numeric, but it can also be a matrix in distributions like the
multivariate normal and the Wishart). The optional argument
prototype can be used to define the default parameter
values in case they are not specified by the user.
Defining a Generator
Now that the class is defined, one can type
D <- new("Beta", shape1 = shape1, shape2 = shape2) to
create a new object of class Beta. However, this is not so
intuitive, and a wrapper function with the class name can be used
instead. This function, often called a generator, can be used
to simply code D <- Beta(1, 2) and define a new object
from the
distribution. The parameter slots can be accessed with the
@ sign, as shown below.
Beta <- function(shape1 = 1, shape2 = 1) {
new("Beta", shape1 = shape1, shape2 = shape2)
}
D <- Beta(1, 2)
D@shape1
D@shape2Defining Validity Checks
This step is optional but rather essential. So far, a user could type
D <- Beta(-1, 2) without any errors, even though the
beta parameters are defined in
.
To prevent such behaviors (that will probably end in bugs further down
the road), the developer is advised to create a
setValidity() function, including all the necessary
restrictions posed by the parameter space.
setValidity("Beta", function(object) {
if(length(object@shape1) != 1) {
stop("shape1 has to be a numeric of length 1")
}
if(object@shape1 <= 0) {
stop("shape1 has to be positive")
}
if(length(object@shape2) != 1) {
stop("shape2 has to be a numeric of length 1")
}
if(object@shape2 <= 0) {
stop("shape2 has to be positive")
}
TRUE
})Defining the Class Methods
Now that everything is set, it is time to define methods for the new
class. Creating functions and S4 methods in R
are two very similar processes, except the latter wraps the function in
setMethod() and specifies a signature class, as shown
below. The package source code can be used to easily define all methods
of interest for the new distribution class.
# probability density function
setMethod("d", signature = c(distr = "Beta", x = "numeric"),
function(distr, x) {
dbeta(x, shape1 = distr@shape1, shape2 = distr@shape2)
})
# (theoretical) expectation
setMethod("mean",
signature = c(x = "Beta"),
definition = function(x) {
x@shape1 / (x@shape1 + x@shape2)
})
# moment estimator
setMethod("me",
signature = c(distr = "Beta", x = "numeric"),
definition = function(distr, x) {
m <- mean(x)
m2 <- mean(x ^ 2)
d <- (m - m2) / (m2 - m ^ 2)
c(shape1 = d * m, shape2 = d * (1 - m))
})